


DNA-Probe for Non-Destructive Chromatin Sequence Extraction (nodeChrose) human genetics, chromatin, purification, DNA, LNA, nucleic acid, DNA probe, oligonucleotide, single-molecule, sequencing
human genetics, chromatin, purification, DNA, LNA, nucleic acid, DNA probe, oligonucleotide, single-molecule, sequencing
DNA-Probe for Non-Destructive Chromatin Sequence Extraction (nodeChrose)
Background:
DNA in eukaryotic cells is folded into chromatin, i.e. every 200 base pairs of DNA wrap around a core of histone proteins forming nucleosomes. Both the composition and the location of the nucleosomes play decisive roles in determining the organization of the whole chromatin complex. Moreover, differences in the regulation of the genes encoded in the DNA have been attributed to different chromatin configurations, giving organisms a means to activate specific sets of genes producing a selected set of proteins in different organs, while maintaining identical DNA copies in all cells. In fact, any intervention in the regulation of transcription, including activation/silencing of genes, involves not just bare DNA, but the complex of DNA and histone proteins.
Nucleosomes form a highly variant class. Their specific variable features include their positioning on DNA along the double-helix, and the occurrence of a number of post-translational modifications on DNA and histones. The occurrence of post-translational modifications is highly regulated and different characters are found in different organs. If fact, misregulation of post-translational modifications can be the origin of (epigenetic) diseases that can even be transferred from generation to generation.
The treatment of the (epi-)genetic diseases might greatly benefit from the capability of monitoring and/or influencing the positions and modifications of histones in chromatin. To avail of selected chromatin fragments extracted from the cell with their intact histone endowment and chromatin structure, is one key to the success of epigenetics research.
Technology Overview:
This technology can select and “pull down” sequence-specific chromatin fragments in a non-destructive way. This allows for highly focused analysis. i.e. zooming in on a single gene, of DNA sequences with their intact histone protein endowment.
At the core of this technology is a novel DNA-probe oligomer formulation, and a methodology to use it for efficient non-destructive chromatin sequence extraction (nodeChrose). This formulation returns a high-affinity probe which is specific to chromatin fragments embedding a known DNA sequence, the “target”, which is long enough to be unique in the genome.
In more detail, the probe is an especially designed oligonucleotide with a target-binding sequence at one end, in which some bases are LNA nucleotides (see “further details”, point (a) ), and at the other end is a covalently bound biotin, which selectively binds to streptavidin coated magnetic beads, allowing an easy pull-down of the extracted chromatin fragments (see “further details”, point (b) ).
The extraction process is initiated by the action of suitable restriction enzymes which selectively cut the chromatin chain and expose a short single stranded DNA portion, the “toehold”, where the oligonucleotide probe can at first “land” and bind. Thereafter, a “strand invasion” occurs at the cleaved end of the chromatin fragment (see figure) allowed by the transient opening of the double stranded DNA and promoted by the high affinity of the LNA-modified nucleotides at the oligonucleotide’s end.
Most remarkably, the whole nodeChrose process happens at room temperature. Such conditions are permissive for the purification of DNA-protein complexes under “native conditions”, where protein-DNA complexes are interacting as they are within a living cell, without the need for crosslinking agents, and without damage to the chromatin fragment nucleosome structure.
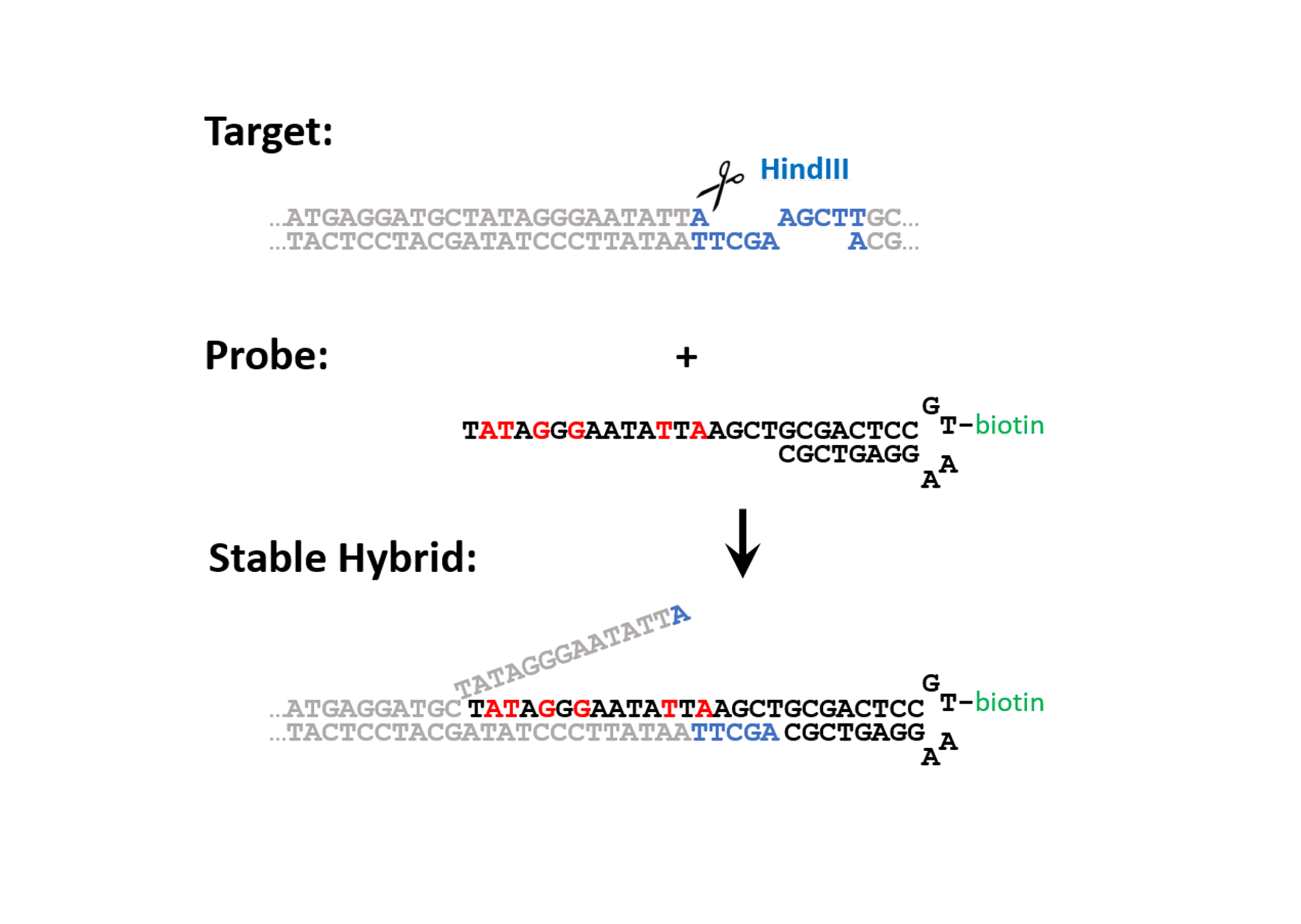
Figure 1: Design of the probe and generalized mechanism for the invasion of the probe into the target. The target is cut with a restriction enzyme creating DNA-toehold of 4 unpaired nucleotides. The probe consists of an 18 base pair overhang (complementary to the target sequence), a DNA hairpin where a modified base can be incorporated (e.g. a biotin), and a stacking sequence that caps the open end of the target. To increase the affinity of the overhang of the probe to the target sequence contains six LNA bases (colored in red). The mechanism of strand invasion can be summarized in four steps: 1) An endonuclease cleaves the target sequence such that a toehold appears. 2) The probe binds the target at the toehold. 3) Fraying of the double stranded target DNA adjacent to the probe drives strand invasion. 4) A stable hybrid between the probe and the target is acquired, which can be further purified by affinity purification.
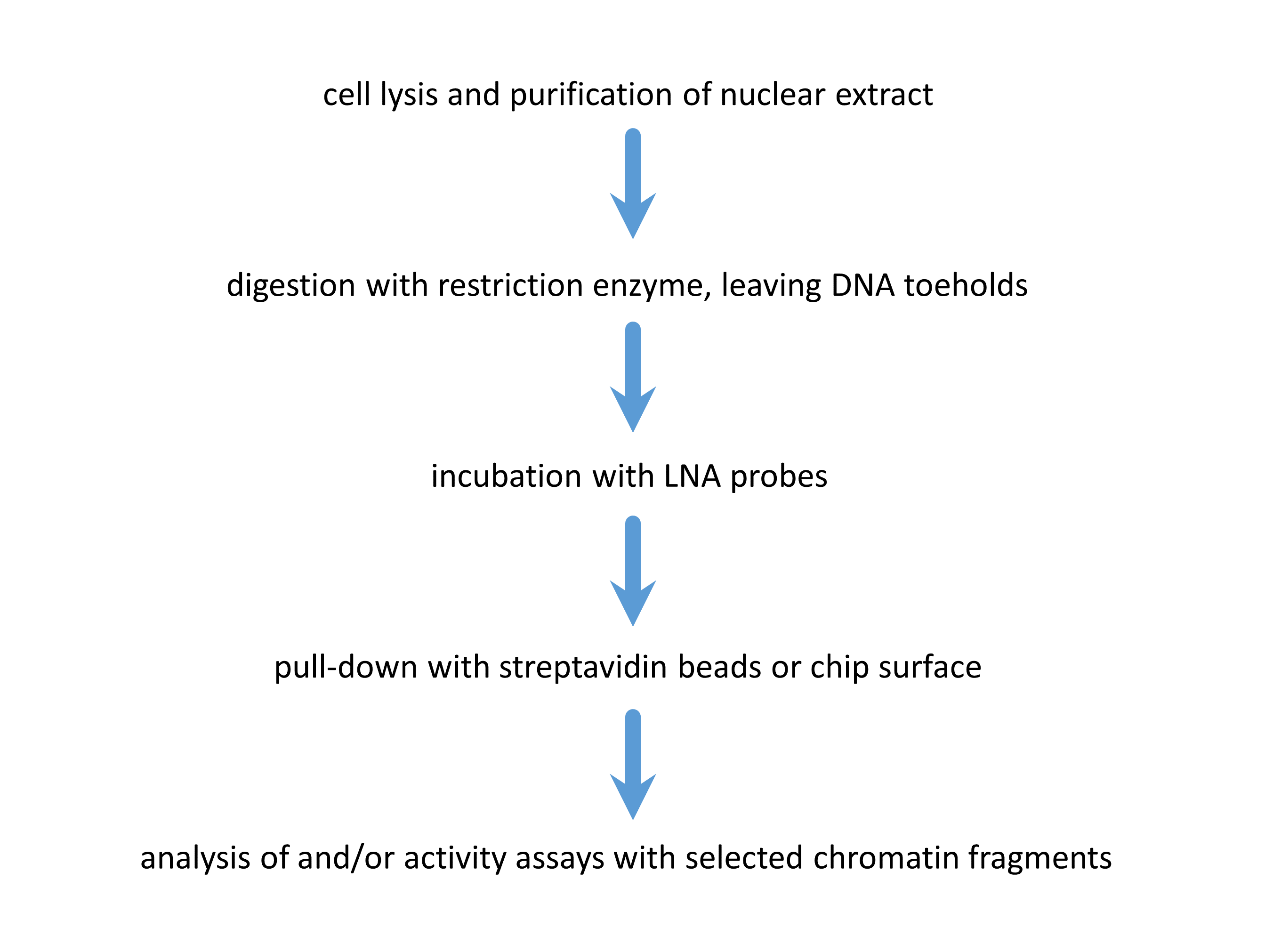
Figure 2: Experimental flow. First, the sample containing the DNA of interest is cut with a restriction enzyme, creating the toehold. Subsequently, the sequence specific probe containing the ligand for immobilization is added, so probe-target hybridization can occur. After this, the target can be pulled down with magnetic beads, or alternatively, immobilized on a surface for further analysis.
Benefits:
This technique overcomes most drawbacks of the other available methods for the extraction of nucleic acids, in that the latter employ any of the following:
- The use of high temperatures (80°C) to separate the two DNA strands and to allow oligonucleotide probes to clasp a target DNA sequence. While efficient, these methods denature completely the histone proteins and dissociate them from the DNA, with a total loss of all epigenetic features.
- The artificial creation of nucleosome structures in extracted DNA sequences. Although useful under many respects, these methods do not provide any specific epigenetic information, in that the obtained artificial chromatin does not reflect the chromatin in the living cells.
- Chemical cross-linking. This keeps the proteins attached to each other and the DNA, but inhibits further activity of analysis as the proteins are chemically different/non-functional. Moreover, chemical crosslinking is known to introduce artefacts.
- Bulk, averaged analysis, which obviously hides variations between chromatin compositions.
Further Details:
(a) Locked nucleic acids (LNA) residues are incorporated to increase the affinity of the probe for the target. LNA nucleotides are modified RNA nucleotides, with an extra covalent bond between the 2' oxygen and the 4' carbon. This modification results in a greater stability of the conformation of the sugar that favours hybridization, and results in higher melting temperatures for duplexes containing LNA bases.
(b) A sequence-specific purification of nucleic acids can be performed with a probe containing a biotin and magnetic beads coated with streptavidin. Furthermore, the target DNA molecule can be linked to two probes, one on each side, to increase specificity. A purification of the target can be done by, e.g., a pull down with magnetic beads, and/or by immobilisation on a surface of proteins with high affinity with the ligand attached to the probe (e.g. streptavidin).
Potential Applications:
This method can be applied in the field of protein research, as well as capturing DNA with a chip based approach in high throughput.
Single molecule analysis of native chromatin, as allowed by this technology, would be particularly useful for:
- any scientific research addressing epigenetics: it is highly cost-effective and has unprecedented resolution
- diagnosis: what does the chromatin landscape look like for a particular gene? Variations in chromatin composition have been linked to a variety of diseases.
- drug lead discovery in the field of epigenetics: once able to purify native chromatin fragments, one has the proper substrate for epigenetic enhancer/silencing factors. These could be used to identify compounds that interfere with such tasks.
State of Development:
This technology was originally developed to enable the use of single-molecule Force Spectroscopy on specific fragments of folded DNA/Histone complexes (chromatin), and as such it was already successfully employed, yielding insight on chromatin’s protein content and characteristics (this is reported in the upcoming scientific publication).
Opportunity:
This technology is already available for exclusive and non-exclusive licensing for commercial use, or for evaluation. However, should relevant opportunities for co-development present themselves, they would be surely taken into consideration, too.
Plasma biomarker for detection of onset of chronic Rheumatoid Arthritis Rheumatoid Arthritis, biomarker, plasma
Rheumatoid Arthritis, biomarker, plasma
Plasma biomarker for detection of onset of chronic Rheumatoid Arthritis
Background:
Rheumatoid Arthritis is characterized by inflammation of joints resulting in joint damage and disability. Research demonstrated that the presence of Rheumatoid Arthritis specific auto-antibodies directed against citrullinated proteins (ACPA) in the serum of patients increased the risk of developing the disease in patients with pain in their joints. ACPA can already be detected in patients years before onset of disease.
Technology Overview:
Researchers at the LUMC have recently discovered that ACPA isolated from Rheumatoid Arthritis patients is decorated with unique sugar structures. These sugar structures are not, or to a lesser extent, present on ACPA of people not yet diagnosed with Rheumatoid Arthritis. This unique sugar structure attached to ACPA could represent a biomarker of the transition phase from healthy to disease. Currently, researchers at the LUMC are investigating the possibility of a diagnostic test to analyse the sugar structures attached to ACPA on a large scale. Based on this test, the sugar structures of ACPA can predict the development of Rheumatoid Arthritis in patients with joint pain. This knowledge is important for clinicians to select an appropriate treatment in time to prevent progression towards chronic Rheumatoid Arthritis.
%2Bfigure%2B1.jpg) [Figure 1: ACPA with unique sugar structures are specifically present in patients with Rheumatoid Arthritis and might be crucial to predict progression from auto-antibody positive healthy subjects to patients with full-blown, chronic and persistent arthritis.]
[Figure 1: ACPA with unique sugar structures are specifically present in patients with Rheumatoid Arthritis and might be crucial to predict progression from auto-antibody positive healthy subjects to patients with full-blown, chronic and persistent arthritis.]
Benefits:
ACPA appear in the blood of Rheumatoid Arthritis patients up to ten years before onset of disease. Currently it is impossible to predict the exact time point of disease manifestation. Therefore, treatment starts at diagnosis of the disease. At this stage, the disease is already chronic, and the patient is devoted to lifelong treatment. This invention allows clinicians to intersect the healthy auto-reactive positive pre-disease phase from the pathogenic phase in which Rheumatoid Arthritis development commenced. If in this stage therapy is applied, development towards Rheumatoid Arthritis might be stopped before chronification occurs.
Further Details:
More background information can be found in the publication: “Extensive glycosylation of ACPA-IgG variable domains modulates binding to citrullinated antigens in rheumatoid arthritis.”, Rombouts et al., Ann Rheum Dis. 2016 Mar.
Potential Applications:
Identify ACPA+ individuals with high risk to develop Rheumatoid ArthritisTargeted treatment - distinguish individuals with high risk to develop Rheumatoid Arthritis in the “at risk” group and start treatmentEarlier start of treatment to prevent/delay/decrease severity and chronification of Rheumatoid Arthritis
Opportunity:
The researchers are looking for partner(s) to license and further (co-)develop and market this assay for clinical application. Specifically companies that are developing diagnostic tools for inflammatory diseases.
IP Status:
Priority patent filed
Novel automated sample purification and enrichment for DI-MS analytical (bio)chemistry, mass spectroscopy
analytical (bio)chemistry, mass spectroscopy
Novel automated sample purification and enrichment for DI-MS
Scientists at Leiden University have invented a method for easy and automated (bio)sample preparation for DI-MS.
The method allows for easy and fast sample purification and enrichment and can easily be integrated with commercially available nanoESI robots.
The aim of Direct-Infusion Mass Spectrometry (DI-MS) is to provide compositional information as well as identification and quantification of specific analytes. Especially nanoelectrospray (nanoESI) is very powerful, but nanoESI emitters are susceptible to clogging due to e.g. protein precipitation and salt crystallization.
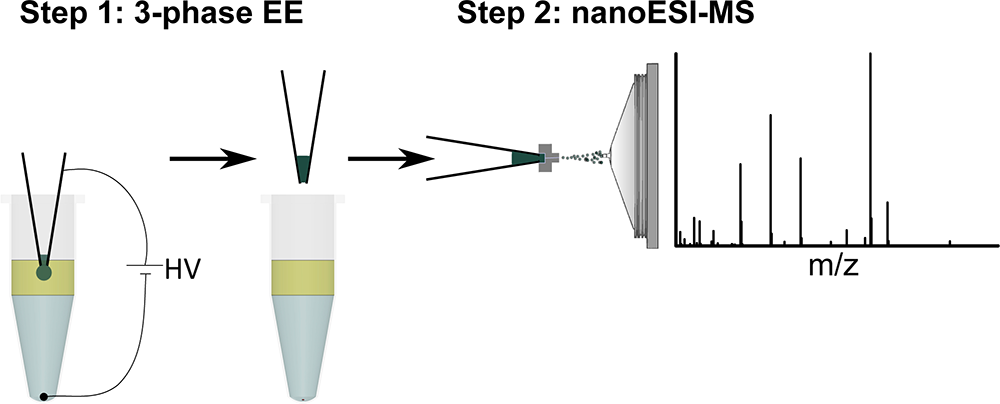
The new preparation method developed in Leiden is based on electroextraction and can be used to selectively extract cations or anions from a sample before infusion into the Mass Spectrometer. This way, for example, proteins and salts can be discarded and therefore the quantitative power of DI-MS can be significantly improved. The method also has the potential to pre-concentrate analytes before infusion into the MS. This entire method has the additional advantage of being easily integrateble with current (robotic) liquid handling/nanoESI systems.
Leiden University is looking for partners to bring this technology to the market.
Type 1 Diabetes Neo-Epitopes: Autoimmunity Against a Defective Ribosomal Insulin Gene Product A new neo-epitope generated by defective ribosomal products (DRiPs) from the proinsulin gene.
A new neo-epitope generated by defective ribosomal products (DRiPs) from the proinsulin gene.
Type 1 Diabetes Neo-Epitopes: Autoimmunity Against a Defective Ribosomal Insulin Gene Product
Background
Type 1 diabetes (T1D) is an autoimmune disease where the immune system destroys the insulin-producing pancreatic beta cells. These cells are insulin factories dedicated to the maintenance of glucose homeostasis; insulin, stored in secretory granules, represents 10–15% of the protein content of these cells. Studies of samples from humans with T1D and mouse models of the disease indicate that native insulin and its precursors act as primary autoantigens, and fragments of the preproinsulin peptide have been identified as main targets of cytotoxic islet-autoreactive CD8+ T cells in human T1D. In autoimmune disease, there is increasing evidence that local inflammation or other forms of stress combined with genetic disposition leads to the generation and the accumulation of aberrant or modified proteins (neo-epitopes) to which central tolerance is lacking and thereby triggering autoimmunity. Until now, neo-epitopes have been shown to be generated through transcriptional, post-transcriptional and post-translational processes. However, an important class of neo-epitopes could be generated through non-conventional translational events and this technology is the first evidence of a naturally processed and presented epitope derived from nonconventional islet proteins.
Technology Overview
Scientists at Leiden University Medical Center have recently discovered a new neo-epitope generated by defective ribosomal products (DRiPs) from the proinsulin gene. Within the mRNA, the researchers have found an alternative open reading frame encoding a highly immunogenic polypeptide that is targeted by T cells in type 1 diabetes (T1D) patients. They show that cytotoxic T cells directed against the N-terminal peptide of this nonconventional product are present in the circulation of individuals diagnosed with T1D, and provide direct evidence that such CD8+ T cells are capable of killing human beta cells and thereby may be diabetogenic. This is the first evidence of a naturally processed and presented epitope derived from nonconventional islet proteins leading to the destruction of human beta cells by cytotoxic CD8+ T cells. They propose a new pathway of beta cell destruction by the immune system in which the generation of a neoepitope, such as INS-DRiP, plays a central role. Their findings support the emerging concept that beta cells are destroyed in T1D by a mechanism comparable to classical antitumour responses whereby the immune system has been trained to survey for dysfunctional cells in which errors have accumulated. This invention reveals a potential new mechanism underlying the pathology of T1D, and may allow the development of novel T1D diagnostics and therapies (Nat Med. 2017 Apr;23(4):501-507).
Details and State of Development:
Proof of concept.
Applications
- Novel T1D diagnostics
- Novel T1D therapies
Opportunity
- Research collaboration to further unravel the mechanism of INS-DriP formation
- Development of novel T1D diagnostics and therapies
DNA-Probe for Non-Destructive Chromatin Sequence Extraction (nodeChrose) This technology can select and “pull down” sequence-specific chromatin fragments using a non-destructive, non-denaturing method
This technology can select and “pull down” sequence-specific chromatin fragments using a non-destructive, non-denaturing method
DNA-Probe for Non-Destructive Chromatin Sequence Extraction (nodeChrose)
Background
DNA in eukaryotic cells is folded into chromatin, ie every 200 base pairs or DNA wrap around a core or histone proteins forming nucleosomes. Both the composition and the location of the nucleosomes play decisive roles in determining the organization of the whole chromatin complex. Moreover, differences in the regulation of the genes encoded in the DNA have been attributed to different chromatin configurations, giving organisms a means to activate specific sets or genes producing a selected set of proteins in different organs, while maintaining identical DNA copies in all cells. In fact, any intervention in the regulation of transcription, including activation / silencing or genes, involves not just DNA, but the complex or DNA and histone proteins.
Nucleosomes form a highly variant class. Their specific variable features include their positioning on DNA along the double-helix, and the occurrence of a number of post-translational modifications on DNA and histones. The occurrence of post-translational modifications is highly regulated and different characters are found in different organs. If fact, misregulation of post-translational modifications can be the origin of (epigenetic) diseases that can even be transferred from generation to generation.
The treatment of the (epi-)genetic diseases might greatly benefit from the capability of monitoring and/or influencing the positions and modifications of histones in chromatin. To avail of selected chromatin fragments extracted from the cell with their intact histone endowment and chromatin structure, is one key to the success of epigenetics research
Technology Overview
This technology can select and “pull down” sequence-specific chromatin fragments in a non-destructive way. This allows for highly focused analysis. i.e. zooming in on a single gene, of DNA sequences with their intact histone protein endowment. At the core of this technology is a novel DNA-probe oligomer formulation, and a methodology to use it for efficient non-destructive chromatin sequence extraction (nodeChrose). This formulation returns a high-affinity probe which is specific to chromatin fragments embedding a known DNA sequence, the “target”, which is long enough to be unique in the genome (Figure 1).
In more detail, the probe is an especially designed oligonucleotide with a target-binding sequence at one end, in which some bases are LNA nucleotides (see “further details”, point (a) ), and at the other end is a covalently bound biotin, which selectively binds to streptavidin coated magnetic beads, allowing an easy pull-down of the extracted chromatin fragments (see “further details”, point (b) ).
The extraction process is initiated by the action of suitable restriction enzymes which selectively cut the chromatin chain and expose a short single stranded DNA portion, the “toehold”, where the oligonucleotide probe can at first “land” and bind. Thereafter, a “strand invasion” occurs at the cleaved end of the chromatin fragment (see figure) allowed by the transient opening of the double stranded DNA and promoted by the high affinity of the LNA-modified nucleotides at the oligonucleotide’s end (Figure 2).
Most remarkably, the whole nodeChrose process happens at room temperature. Such conditions are permissive for the purification of DNA-protein complexes under “native conditions”, where protein-DNA complexes are interacting as they are within a living cell, without the need for crosslinking agents, and without damage to the chromatin fragment nucleosome structure.
Details and State of Development:
- This technology was originally developed to enable the use of single-molecule Force Spectroscopy on specific fragments of folded DNA/Histone complexes (chromatin), and as such it was already successfully employed, yielding insight on chromatin’s protein content and characteristics (this is reported in the upcoming scientific publication).

Figure 1: Design of the probe and generalized mechanism for the invasion of the probe into the target. The target is cut with a restriction enzyme creating DNA-toehold of 4 unpaired nucleotides. The probe consists of an 18 base pair overhang (complementary to the target sequence), a DNA hairpin where a modified base can be incorporated (e.g. a biotin), and a stacking sequence that caps the open end of the target. To increase the affinity of the overhang of the probe to the target sequence contains six LNA bases (colored in red). The mechanism of strand invasion can be summarized in four steps: 1) An endonuclease cleaves the target sequence such that a toehold appears. 2) The probe binds the target at the toehold. 3) Fraying of the double stranded target DNA adjacent to the probe drives strand invasion. 4) A stable hybrid between the probe and the target is acquired, which can be further purified by affinity purification.

Figure 2: Experimental flow. First, the sample containing the DNA of interest is cut with a restriction enzyme, creating the toehold. Subsequently, the sequence specific probe containing the ligand for immobilization is added, so probe-target hybridization can occur. After this, the target can be pulled down with magnetic beads, or alternatively, immobilized on a surface for further analysis.
Applications
1. Any scientific research addressing epigenetics: it is highly cost-effective and has unprecedented resolution
2. Diagnosis: what does the chromatin landscape look like for a particular gene? Variations in chromatin composition have been linked to a variety of diseases.
3. Drug lead discovery in the field of epigenetics: once able to purify native chromatin fragments, one has the proper substrate for epigenetic enhancer/silencing factors. These could be used to identify compounds that interfere with such tasks.
Opportunity
This technology is already available for exclusive and non-exclusive licensing for commercial use, or for evaluation. However, should relevant opportunities for co-development present themselves, they would be surely taken into consideration too.
Keywords: human genetics, chromatin, purification, DNA, LNA, nucleic acid, DNA probe, oligonucleotide, single molecule, sequencing
Tumor Associated Carbohydrate Antigens as Targets for Colorectal Cancer Immunotherapy The present invention relates to a novel method to identify highly specific tumor associated carbohydrate antigens (TACAs) derived from colorectal cancer (CRC) tissue.
The present invention relates to a novel method to identify highly specific tumor associated carbohydrate antigens (TACAs) derived from colorectal cancer (CRC) tissue.
Tumor Associated Carbohydrate Antigens as Targets for Colorectal Cancer Immunotherapy

Summary
The present invention relates to a novel method to identify highly specific tumor associated carbohydrate antigens (TACAs) derived from colorectal cancer (CRC) tissue. Furthermore, it is believed that the markers disclosed may be useful therapeutic targets easily accessible as expressed on cell surfaces directly accessible to therapeutics and can be carried by multiple
proteins, reflecting the overall glycosylation phenotype of the cell, providing a broader tumor targeting strategy.
There was a patent filed recently which covers the workflow of extracting, identifying and analysing these highly specific structures as well as another one that includes the specific TACA structures which were found to be solely present in CRC and not in healthy colon providing a number of promising targets for potential therapy.
Background
Colorectal cancer (CRC) is one of the leading malignancies worldwide with over 900,000 deaths in 20201. Conventional therapeutic strategies include chemotherapy, radiation therapy, and surgery. However, due to poor screening strategies and lack of symptoms in early stages, most cases are detected at an advanced stage, leading to unsuccessful treatment. Unraveling the glycome in cancer is important for the development of immunotherapies for treating solid tumors and the discovery of cancer-specific glycan structures is critical for improving how these cancers are targeted.
Technology
The present invention provides a better understanding of the variation in
O-glycosylation and its association with cell phenotypes. An in-depth
structural O-glycosylation analysis of 26 CRC cell lines was performed.
Additionally, the O-glycosylation signatures of primary cell lines as well as the primary and metastatic colorectal cancers they have been derived from were mapped and compared. Furthermore, glycosylation signatures of paired micro-dissected cancer and healthy mucosa were investigated. The released O-glycans were analysed on a sensitive nano-liquid chromatography coupled to a tandem mass spectrometer using electrospray ionization enabling powerful separation of isomeric species, as well as in-depth structural characterization of the epitopes. The major outcomes were as follows:
- Using transformative technology based on laser capture microdissection allowed for the first time to dissect the cancer cell O-glycome of CRC.
- Highly specific glycan structures were observed in cancer cells from most of the tumor tissue which were not present in healthy control tissue.
Value Proposition
The global colorectal cancer therapeutics market is poised to grow by USD 994.94 million during 2019-2023, progressing at a CAGR of almost 3% (www.businesswire.com).
Team
Prof Manfred Wuhrer currently holds the position as Head of the Center for Proteomics and Metabolomics (CPM) at the Leiden University Medical
Center in Leiden/ The Netherlands. Dr Guinivere Lageveen Kammeijer is
Senior Scientist at the CPM focusing on exploring cancer glycosylation for developing novel therapeutic and diagnostic applications. Dr Katarina
Madunic is Scientist at the CPM specialized in dissecting cancer cell
glycosylation.
Immunomodulatory Capacity and Therapeutic Potential of S.mansoni-Derived Proteins in the Context of Immune Modulation, Asthma and/or Allergy Proteomics analysis on several fractions was performed and identified a number of unique proteins of which some were highly abundant. Identification of unique Breg-inducing molecules may form interesting treatment targets for inflammatory diseases such as asthma or allergies.
Proteomics analysis on several fractions was performed and identified a number of unique proteins of which some were highly abundant. Identification of unique Breg-inducing molecules may form interesting treatment targets for inflammatory diseases such as asthma or allergies.
Immunomodulatory Capacity and Therapeutic Potential of S.mansoni-Derived Proteins in the Context of Immune Modulation, Asthma and/or Allergy

Summary
During chronic schistosome infections, a complex regulatory network is induced to regulate the host immune system, in which IL-10-producing regulatory B cells (Breg) play a significant role. Schistosomal egg antigens (SEA) are bound and internalized by specific murine B cells and induce both human and mouse IL-10 producing Breg cells. To identify Breg inducing proteins in SEA, HPLC-fractionation was applied, showing some active fractions. Proteomics analysis on several fractions was performed and identified a number of unique proteins of which some were highly abundant. Identification of unique Breg-inducing molecules may form interesting treatment targets for inflammatory diseases such as asthma or allergies.
Background
Current treatment of asthma are mainly bronchodilators or inhaled corticosteroids with or without the long-lasting β2-agonists. Daily use of these medications is required, to prevent recurrence of inflammation and bronchoconstriction. In addition to the high therapy compliance, other disadvantages of the commonly used treatment for asthma are possible long-term side effects and the lack of responsiveness in severe, therapy-resistant asthma patients. Certain environmental and health factors increase risk of wheezing or asthma in children, or the pathology of existing asthma. Additionally, neither cure nor preventional therapy for asthma exists, only medication to alleviate symptoms. Avenues that focus on primary prevention have the potential to open a completely new market. Helminths derived molecules represent an attractive source of new immunomodulatory therapeutics/biologicals for use in asthma (or allergy) treatment.
Technology
Some candidate proteins were identified as potentially immunomodulatory in fractions of parasite material with the capacity to induce IL-10 from mouse B cells. These crude molecules also seem to be able to block type 2 innate cytokines from human primary bronchial epithelial cells. They were therefore recombinantly produced using specific separation techniques. One specific candidate molecule has also
been tested against human B cells, whereby it also shows the capacity to
induce IL-10. In addition, these pure molecules will also be tested in bronchial epithelial cells to evaluate their capacity to block respiratory infections, such as rhinoviruses.
Value Proposition
The global Asthma and COPD Drugs Market Size alone was valued at
$32988.7 million in 2020 and is projected to reach $52049.54 million by
2030, registering a CAGR of 4.64% from 2021 to 2030. Asthma is a noncommunicable, chronic inflammatory lung disorder of the airways
(www.alliedmarketresearch.com).
Team
Prof. Hermelijn H. Smits currently holds the position as Professor in 'Host-commensal interactions and Immunemodulation' at the Parasitology department within the Leiden University Medical Center in
Leiden/ The Netherlands. She worked at this invention together with her
team.

